Nanopore’s yottasteps
| 20 March, 2017 | Hollydawn Murray |
|

|
Hollydawn Murray, gives us a run down of recent events in the Nanopore field and speaks to F1000Research author Scott Gigante who has been part of the recent excitement
Credit: Jonathan Bailey, NHGRI
Between PoreCamp Australia and the Advances in Genome Biology and Technology (AGBT) General Meeting, February was an exciting month for Nanopore and it hasn’t ended there. Last week, Clive Brown of Oxford Nanopore Technologies delivered an impressive technical update: GridION X5 – The Sequel. When asked ‘how can I prepare ~1 Mb size DNA?’, Clive advised ‘be gentle’. This question likely stems from the 771
kilobase ultra-long read recently achieved by Nick Loman and Josh Quick of the University of Birmingham; that’s a whopping 1/6th of the E.coli genome sequenced in a single read! In a blog detailing the full protocol, Nick claimed four world records (longest mappable DNA strand sequence, longest mappable DNA strand & complement sequence, highest Nanopore run N50, and highest Nanopore run mean read length) and predicted that the megabase read is in sight. And Nick is not alone: Keith Robison, of the Omics! Omics! blog, said he would be surprised if a megabase read isn’t reported at or before London Calling in May.
And while read length has become a focus of the Nanopore community, it’s not the only size that matters; increases in read length, 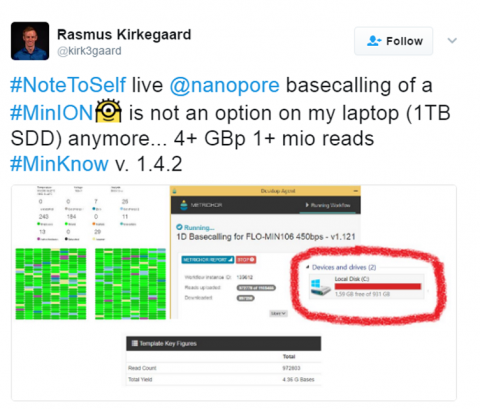 basecalling accuracy, and total number of reads yield increasingly large datasets. Such large datasets pose space issues for both immediate and long-term data storage. Enter ‘Picopore’, a tool for reducing the size of Nanopore datasets, conceptualised and developed by Scott Gigante during PoreCamp Australia. The paper is currently awaiting peer review (Update, 21/3/2017: the article has now passed peer review). Scott spoke to me about his experience, and Picopore in more detail.
basecalling accuracy, and total number of reads yield increasingly large datasets. Such large datasets pose space issues for both immediate and long-term data storage. Enter ‘Picopore’, a tool for reducing the size of Nanopore datasets, conceptualised and developed by Scott Gigante during PoreCamp Australia. The paper is currently awaiting peer review (Update, 21/3/2017: the article has now passed peer review). Scott spoke to me about his experience, and Picopore in more detail.
What was your first experience with Nanopore?
 I started working with Nanopore data in June last year, when I started at the Walter & Eliza Hall Institute (WEHI) as an undergraduate research student. My project was to use recurrent neural networks to predict base modifications on Nanopore reads. I’ve since graduated and taken up a full time research role at WEHI, still working primarily with Nanopore data. However, despite working on the data for nine months now, my first experience actually using the MinION in the lab was just a month ago, as part of PorecampAU!
I started working with Nanopore data in June last year, when I started at the Walter & Eliza Hall Institute (WEHI) as an undergraduate research student. My project was to use recurrent neural networks to predict base modifications on Nanopore reads. I’ve since graduated and taken up a full time research role at WEHI, still working primarily with Nanopore data. However, despite working on the data for nine months now, my first experience actually using the MinION in the lab was just a month ago, as part of PorecampAU!
Why did you create Picopore, and how did you approach it?
One of the major lessons we learned as participants of PorecampAU was that the hard drive specifications given by Oxford Nanopore Technologies (ONT) are really an absolute minimum, not an optimum. This was very much learned the hard way; between the ten or so of us who were sitting at or below specification, we must have lost over a million reads due to a lack of storage space! From my point of view, requiring users to own a terabyte solid state drive in order to use the MinION was a significant barrier to the portability and accessibility of the MinION, which is otherwise revolutionary in these regards.
I had experience manipulating FAST5 files from my previous projects, and therefore had an idea of where some savings could be made. Further research into the file format showed me additional methods of improvement. Having analysed these files as both a developer and an end-user, I also had an idea of what data had to be retained in order to maintain their usability; from here I devised the three methods of compression that make up Picopore. By lunch on the third day of Porecamp, I had begun programming the solution.
What was the highlight of your Porecamp experience?
The highlight of my Porecamp experience, as mundane as this might sound, was to actually prepare the DNA and load it into the MinION. I’m a mathematician and computer scientist by training; prior to Porecamp, the last time I held a pipette was in high school! It was a rewarding learning experience to see first-hand the process of preparing the DNA, and to understand the work and finesse that goes into preparing the files that I have been manipulating for the last nine months.
You tweeted that Picopore went from concept to publication in just 26 days. Where do you find the energy, and what’s next?
As anyone watching the Nanopore community over the last few years would know, it’s a very fast-moving area. What is a good idea today is irrelevant tomorrow, so I knew I had to be on my toes. That said, in all honestly it was a bit of an accident that Picopore happened so quickly!
I started work on Picopore, as I do with any project, with a standard (public) GitHub repository. At some stage while I was testing a few things on the Saturday after Porecamp (less than 36 hours after I had the idea) somebody discovered my repository on GitHub and shared it on Twitter. This was a call to action – I couldn’t have a half-done program circulating around the community! I pushed myself to complete an alpha release by that Sunday night. The remaining 24 days were mostly spent polishing the code base, and writing up the results for publication. Of course I couldn’t have published so quickly without the innovative format of F1000Research, for which I am very grateful!
As for what’s next: if we follow the model of Picopore, who knows? I’m very interested in machine learning, and I see a number of opportunities in improving and expanding Nanopore basecalling using different neural network architectures. Also, considering the pace of development in Nanopore, particularly with the “leviathan” reads coming out of Nick Loman’s lab at the University of Birmingham, it is clear that there is a lot of work to be done in improving downstream analysis for ultra-long reads. There’s certainly more to be done in this space than any one person can take on; it just depends what jumps out at me next!

|



User comments must be in English, comprehensible and relevant to the post under discussion. We reserve the right to remove any comments that we consider to be inappropriate, offensive or otherwise in breach of the User Comment Terms and Conditions. Commenters must not use a comment for personal attacks.
Click here to post comment and indicate that you accept the Commenting Terms and Conditions.