Addressing the (in)consistency of pharmacogenomic datasets
| 13 February, 2017 | Benjamin Haibe-Kains |
|

|

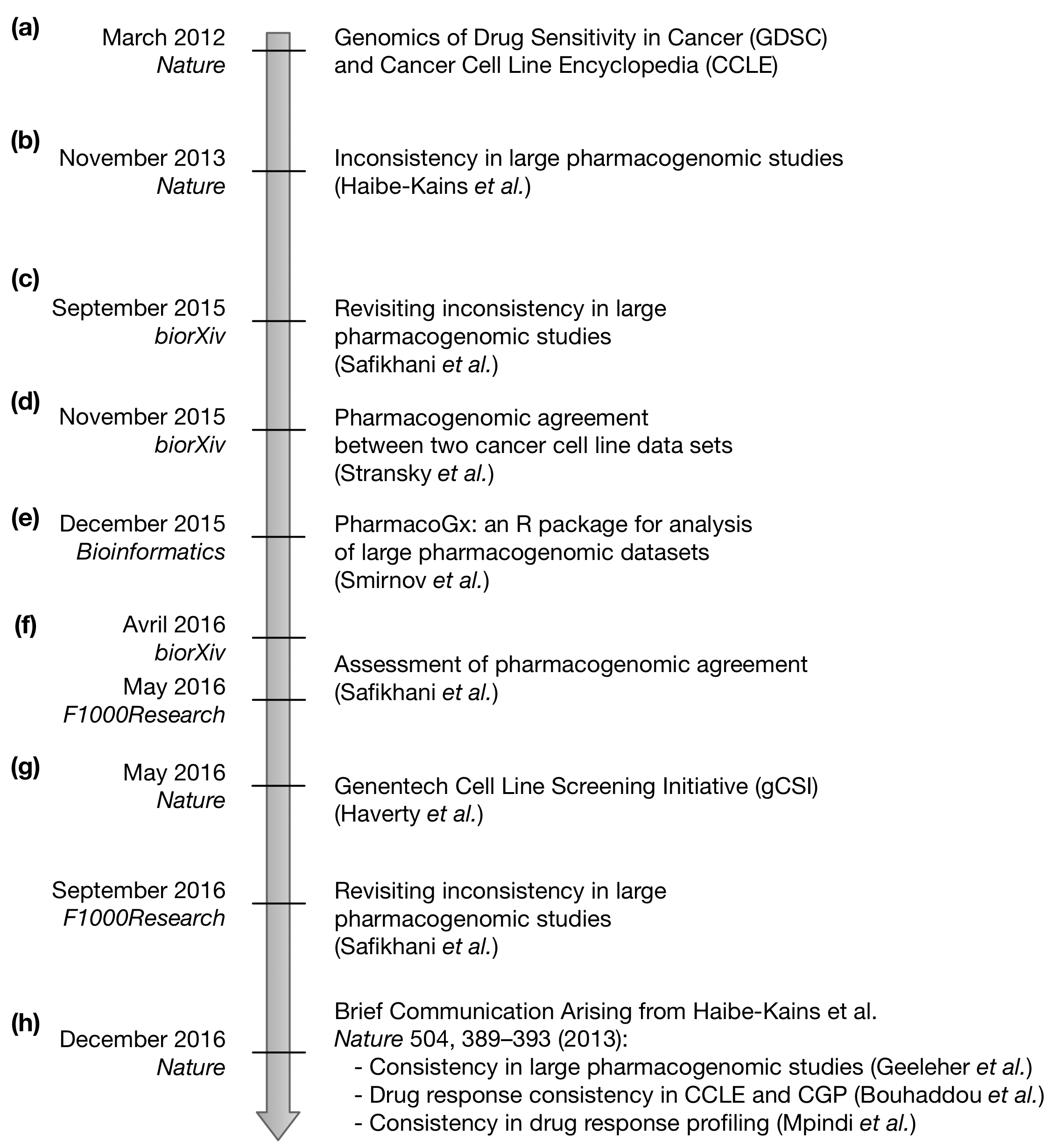
In March 2012, two seminal studies published large pharmacogenomic datasets with drugs screened in hundreds of immortalized cancer cell lines: the Genomics of Drug Sensitivity in Cancer (GDSC) and the Cancer Cell Line Encyclopedia (CCLE) (Figure 1a at the end of the post). These two studies opened new avenues of research for building and testing molecular predictors of drug response. However, we and a couple of other studies showed that using one dataset to find new biomarkers of response and validate their predictive value in the other dataset was challenging. Our subsequent investigation led us to discover inconsistencies between the drug sensitivities data published in the GDSC and CCLE studies (Figure 1b). While gene expression data were reasonably in agreement, we found that the drug sensitivity measures – the drug concentration required to inhibit 50% of cell viability [IC50] and the area under the drug dose-response curve [AUC] – were inconsistent for most of the drugs. Importantly, we showed that these inconsistencies had a negative impact on the development of molecular predictors of drug response.
Revisiting inconsistency in large pharmacogenomic studies
Our comparative study triggered intense discussions in the field and we received extensive feedback on our analyses and results from the community. This feedback, along with the release of new data, prompted us to revisit our initial analysis, which we made available on biorXiv (Figure 1c). In this new study, we addressed important criticisms. These include the lack of discrimination between highly targeted drugs and those with broad effect in our initial study prevented us from reporting the (in)consistency of pharmacological profiles within different contexts, where either drugs have no or little effect except in a small subset of cell lines, or drugs yield a wide spectrum of growth inhibition in the majority of cell lines. However, the conclusions of our revisited analysis were virtually identical to our initial publication: as long as there is a lack of standardisation in experimental assays and analysis methods, users of pharmacogenomics data sets should be cautious in their interpretation of results derived from their analyses and particularly wary about making predictions of clinical response based on in vitro measurements.
Pharmacogenomic agreement between two cancer cell line data sets
In the meantime the GDSC/CCLE investigators published their own comparative study, reporting pharmacogenomic agreement between the GDSC and CCLE datasets (Figure 1d). In order to scrutinize these new exciting results, we leveraged our PharmacoGx platform (Figure 1e) to investigate how two studies comparing the same datasets reached seemingly opposite conclusions. In addition to the usual differences in qualitative interpretation of consistency measurements [is a correlation coefficient ~0.5 enough to claim consistency of drug sensitivity data? Which criteria should be used to claim consistency of biomarkers?] we found that the GDSC/CCLE investigators used an experimental design that fundamentally differed from our initial study. While we analyzed independently the GDSC and CCLE datasets to ensure unbiased data comparison, the GDSC and CCLE investigators used different drug sensitivity data (IC50 or AUC) in their multiple data comparisons and selected a single dataset as the source of molecular profiles, therefore ignoring the noise present in these complex data. We therefore concluded that these analytical choices contributed at least partially to the discrepancy between the two published comparative studies. We made our manuscript available on biorXiv (Figure 1f) to clearly describe the different analytical designs used in the 2013 and 2015 comparative studies.
Reproducible pharmacogenomic profiling of cancer cell line panels
Shortly after, Genentech published a new comparative study using their own data as reference (Genentech Cell Line Screening Initiative [gCSI]; Figure 1g) and found that their drug sensitivity data was more similar to CCLE (which used the same pharmacological assay) than GDSC. This was the only study evaluating different aspects of the screening protocols that are relevant to measured drug response, including the readout of cell viability, seeding density strategy, and cell culture media conditions. The authors found that differences in media conditions and seeding density contributed to inconsistencies between studies, and that metabolic and DNA-content pharmacological assays exhibit markedly different levels of noise. Although gCSI drug sensitivity data were more correlated with CCLE, none of the datasets formed a clear cluster, indicating a substantial level of of noise in the pharmacological profiles.
Using open peer review to improve the consistency of drug sensitivity data
At this point, we had submitted all our analyses to biorXiv. Publishing confirmatory results and comments regarding published articles can be challenging, to say the least. Fortunately, we were contacted by the F1000Research Editors who had just created the Preclinical Reproducibility and Robustness channel, a perfect fit for our work. Particularly appealing was the open peer review process, allowing us to best revise our work based on the reviewers’ feedback.
Our communication arising from the GDSC/CCLE comparative study was reviewed and approved by three established biostatisticians (Figure 1f) while our revisited study is being revised. Given the complexity of the controversy surrounding the (in)consistency of pharmacogenomic datasets, F1000Research also provides a unique platform for post-publication review as any researchers can post comments describing their own observations and perspective. Our initial comparative analysis triggered a series of new studies aiming at improving the consistency between large pharmacogenomic datasets. The three brief communications arising from our initial comparative study and our responses recently published in Nature (Figure 1h) are good examples of alternative approaches that could be used to increase, although only marginally, the consistency of drug sensitivity data. But there is still a dire need for better integrative analysis frameworks to leverage the plethora of large pharmacogenomic studies available to date. This is just the beginning as in vitro and in vivo drug screening platforms will become more and more important in the context of precision medicine. We believe that preprint servers, as well as open access publishing venues, with transparent pre- and post-publication peer-review, such as F1000Research, will be crucial to ensure proper dissemination of data and analysis results in the active field of cancer pharmacogenomic research.

|


User comments must be in English, comprehensible and relevant to the post under discussion. We reserve the right to remove any comments that we consider to be inappropriate, offensive or otherwise in breach of the User Comment Terms and Conditions. Commenters must not use a comment for personal attacks.
Click here to post comment and indicate that you accept the Commenting Terms and Conditions.