The way to my heart
| 1 March, 2011 | Richard P. Grant |
|

|
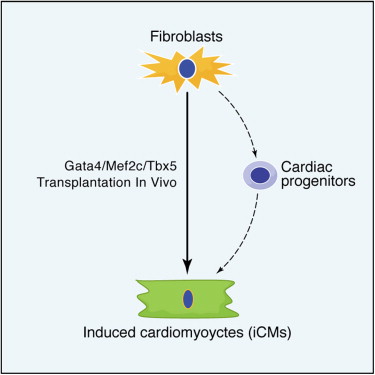
There was a rather wonderful paper in Cell in the middle of last year from Deepak Srivastava’s lab at UCSF: Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors1. It was picked up by two of our Faculty teams and has already garned more than 40 citations.
The core message was that cardiac fibroblasts could be induced to form myocytes directly, without going through an induced pluripotent stem (iPS) cell stage. By a process of elimination, Deepak and his colleagues found that just three transcription factors, Gata4, Mef2c and Tbx5, acting together can induce this reprogramming. Although rather high viral titers (retroviruses or inducible lentiviruses were used to deliver the transcription factor coding sequences) were required to bring this about, transduction efficiency was better than 95% and cultured, infected fibroblasts turned into myocytes within two weeks of transplantation into live mouse hearts.
Unfortunately, my spies tell me that it’s been reported (at a Keystone Conference last month) that other groups are having difficulty reproducing the results. This is a bit of a worry, and does cast a shadow over what could potentially be a way of repairing heart damage after, say, a myocardial infarct. There has been no suggestion of misconduct, and it’s probably down to failures in following published methods: anybody who has tried to follow a method in a paper, especially one that references methods in previous papers, knows how painful that can be.
The groups concerned (say my spies) are working together and sharing reagents and protocols. This does raise the question, however: in the era of internet and cheap data storage, why on earth isn’t it possible to share full methods along with the published paper (Journal of Neuroscience, I’m also looking at you)? One for Peter Murray-Rust to mull over, perhaps.
It’s a little bit more effort on the part of the scientist to begin with (trying to piece together methods from multiple people for a paper was never my favourite task), but in the long run, I can’t see who loses.

|

I agree – and it always seems to be the high impact journals that publish the least amount of methods. It’s a basic scientific writing principle to describe your methods with enough detail such that an expert in the same field could reproduce your results. This was obviously not the case with this publication and points to laxity on the part of reviewers and editors to allow omission of sufficient methodology. The problem has existed for years and I’m glad somebody who might get noticed has finally raised the issue.
Cpme, come. In most labs, the real methods are messy, may not always have worked, and are often incompletely recorded. There can be strange factors involved that nobody knows about which prove important later.
And, of course, there are sometimes fictions… But usually it’s just a matter of incomplete recording of exactly how it was really done.
Not really an excuse, and I agree with the idea. I’m just thinking that the author may be overly optimistic.
I agree with Ellen. We sometimes have trouble getting in situ hybridizations, which have been worked out over fifteen years of tweaking written protocols in our lab, to work from one trainee to another *within our laboratory*. I personally have had the same experience when I was trying to perform SAGE years ago, and more than one group had gotten it to work for them (we did as well, in the end). When others get this to work, there will end up being a consensus protocol that works for more and more people.
I suspect there are fibroblasts and fibroblasts, and differences between strains, that are part of what is going on here. I am quite confident there was no misconduct, though I also lament the rush to publish and not include a detailed protocol in the “best” journals so that others can reproduce and validate the results.
always seems to be the high impact journals that publish the least amount of methods
this does unfortunately seem to be true.
Heather (nice to see you here 🙂 ) and Ellen–why do you think this is? Is it because our methods, or models or materials, just aren’t robust? Does this suggest a fundamental messiness about biological science that should make us all a bit worried? (See ‘decline effect’, &c.)
The fundamental messiness about biological science is stochastic, in my opinion, and doesn’t bother me too much. Except when the n’th in situ doesn’t work, because it feels *inefficient*.
“Decline effect” is all the fashion now, and commendable as an observation except insofar as it gives fodder to proponents of anti-scientific thinking. Not every article describes a transformative discovery. It describes itself an observation under certain circumstances, but any given article’s approach to truth is by the nature of all human endeavor, partial.
I think understanding aspects of nature is difficult. We are part of an impatient time. But it takes time, patience and often, experience and perspective, to make sense in the long run. I am pretty confident that the findings from the Srivastava group will be partially validated and partially re-interpreted over the long run, just as it has been for pluripotent stem cells from bone marrow and all sorts of other once-exciting findings in biology even in recent memory.
Is it such a problem that “truth” is often a matter of context and interpretation and an asymptotic approach is the only one?